2D example: granular column collapse
By Yongjin Choi, Georgia Institute of Technology Website
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
Problem#
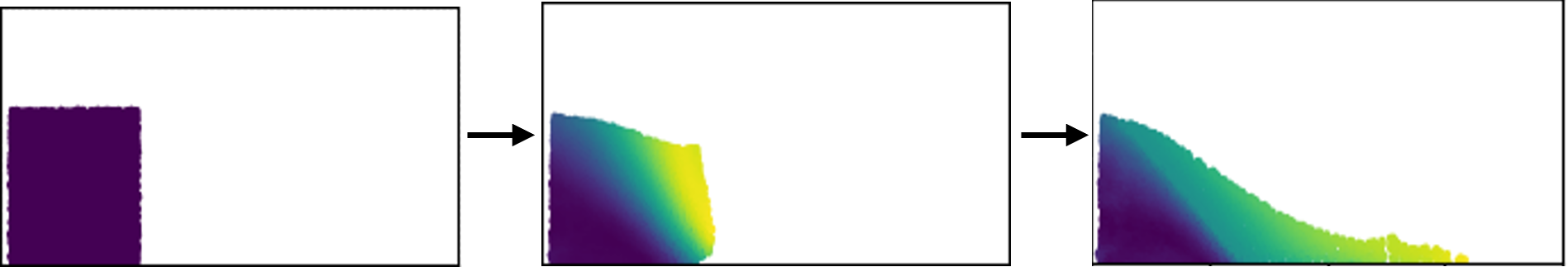
This example considers a granular column collapse problem. The column is represented with material points and then subjected to a collapse due to gravity. The particles are modeled using the Drucker-Prager plasticity model.
Helper functions#
def elasticity_matrix(youngs_modulus: float, poissons_ratio: float, stress_state: str) -> np.ndarray:
"""Compute the elasticity (stiffness) matrix for an isotropic elastic material.
Args:
youngs_modulus: Young's modulus of the material.
poissons_ratio: Poisson's ratio of the material.
stress_state: Type of stress state. Must be one of "PLANE_STRESS",
"PLANE_STRAIN", or "3D".
Returns:
Elasticity matrix as a numpy array.
Raises:
ValueError: If an invalid stress_state is provided.
"""
if stress_state == 'PLANE_STRESS':
C = youngs_modulus / (1 - poissons_ratio**2) * np.array([
[1, poissons_ratio, 0],
[poissons_ratio, 1, 0],
[0, 0, (1 - poissons_ratio) / 2]
])
elif stress_state == 'PLANE_STRAIN':
C = youngs_modulus / ((1 + poissons_ratio) * (1 - 2 * poissons_ratio)) * np.array([
[1 - poissons_ratio, poissons_ratio, 0],
[poissons_ratio, 1 - poissons_ratio, 0],
[0, 0, 0.5 - poissons_ratio]
])
else: # 3D stress state
C = np.zeros((6, 6))
C[:3, :3] = youngs_modulus / ((1 + poissons_ratio) * (1 - 2 * poissons_ratio)) * np.array([
[1 - poissons_ratio, poissons_ratio, poissons_ratio],
[poissons_ratio, 1 - poissons_ratio, poissons_ratio],
[poissons_ratio, poissons_ratio, 1 - poissons_ratio]
])
C[3:, 3:] = youngs_modulus / (2 * (1 + poissons_ratio)) * np.eye(3)
return C
def generate_rectangular_particles(origin, length, height, spacing):
"""
Generate a numpy array that discretizes a rectangular shaped mass into equally spaced material points.
Parameters:
origin (tuple): (x, y) coordinates of the bottom left corner of the rectangle
length (float): Length of the rectangle
width (float): Width of the rectangle
spacing (float): Distance between adjacent particles
Returns:
numpy.ndarray: Array of shape (n, 2) containing the coordinates of material points
"""
# Calculate the number of points along each dimension
nx = int(np.ceil(length / spacing)) + 1
ny = int(np.ceil(height / spacing)) + 1
# Generate equally spaced points along x and y axes
x = np.linspace(origin[0], origin[0] + length, nx)
y = np.linspace(origin[1], origin[1] + height, ny)
# Create a mesh grid
xx, yy = np.meshgrid(x, y)
# Reshape the meshgrid into a 2D array of points
points = np.column_stack((xx.ravel(), yy.ravel()))
return points
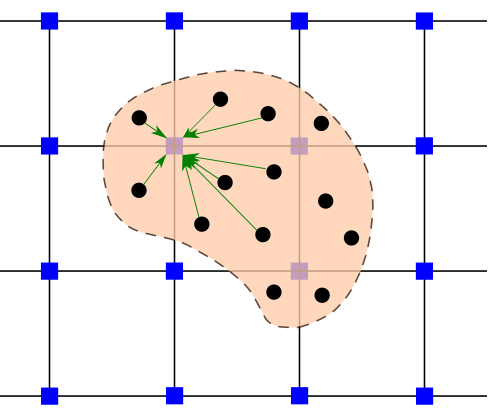
def particle_element_mapping(particle_positions: np.ndarray, cell_width: float, cell_height: float,
n_ele_x: int, n_elements: int):
"""Map each particle to an element based on its position.
Args:
particle_positions: Particle positions as an array of shape (n_particles, 2).
cell_width: Element (cell) width in the x-direction.
cell_height: Element (cell) height in the y-direction.
n_ele_x: Number of elements in the x-direction.
n_elements: Total number of elements.
Returns:
ele_ids: Array of element ids for each particle.
particle_ids_in_elements: List of particle indices for each element.
"""
x_indices = np.floor(particle_positions[:, 0] / cell_width).astype(int)
y_indices = np.floor(particle_positions[:, 1] / cell_height).astype(int)
ele_ids = x_indices + n_ele_x * y_indices
particle_ids_in_elements = [[] for _ in range(n_elements)]
for p, e in enumerate(ele_ids):
particle_ids_in_elements[e].append(p)
return ele_ids, particle_ids_in_elements
def lagrange_basis(element_type: str, local_coord: np.ndarray):
"""Compute the shape functions and their derivatives for a finite element.
Args:
element_type: Type of element. Currently only "Q4" is supported.
local_coord: Array of local coordinates [xi, eta].
Returns:
N: Shape function values.
dN_dxi: Derivatives of the shape functions with respect to xi and eta.
Raises:
ValueError: If an unsupported element type is provided.
"""
if element_type == 'Q4':
xi, eta = local_coord
N = np.array([
(1 - xi) * (1 - eta),
(1 + xi) * (1 - eta),
(1 + xi) * (1 + eta),
(1 - xi) * (1 + eta)
]) / 4.0
dN_dxi = np.array([
[-(1 - eta), -(1 - xi)],
[ (1 - eta), -(1 + xi)],
[ (1 + eta), (1 + xi)],
[-(1 + eta), (1 - xi)]
]) / 4.0
else:
raise ValueError(f"Unsupported element type: {element_type}")
return N, dN_dxi
def compute_local_coord(particle_pos: np.ndarray, element_coords: np.ndarray,
cell_width: float, cell_height: float) -> np.ndarray:
"""Compute the local (ξ, η) coordinates for a particle within an element.
Args:
particle_pos: [x, y] coordinates of the particle.
element_coords: Coordinates of the element nodes (4 x 2 array).
cell_width: Element width in the x-direction.
cell_height: Element height in the y-direction.
Returns:
Local coordinates [ξ, η].
"""
xi = (2 * particle_pos[0] - (element_coords[0, 0] + element_coords[1, 0])) / cell_width
eta = (2 * particle_pos[1] - (element_coords[1, 1] + element_coords[2, 1])) / cell_height
return np.array([xi, eta])
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.cm as cm
# Temporary hard-coded variable
nu = 0.3
def principal_stresses(stress_tensor):
"""
Calculate the principal stresses sigma_1 and sigma_2 for a 2D stress tensor using eigen decomposition.
Parameters:
stress_tensor (list): A list of the form [sigma_xx, sigma_yy, sigma_xy].
Returns:
tuple: Principal stresses (sigma_1, sigma_2) with sigma_1 >= sigma_2.
"""
# Unpack the stress components
sigma_xx, sigma_yy, sigma_xy = stress_tensor
# Form the 2D stress tensor matrix
stress_matrix = np.array([[sigma_xx, sigma_xy],
[sigma_xy, sigma_yy]])
# Perform eigen decomposition to get the eigenvalues (principal stresses)
eigenvalues, _ = np.linalg.eig(stress_matrix)
# Sort eigenvalues in descending order: sigma_1 >= sigma_2
sigma_1, sigma_2 = np.sort(eigenvalues)[::-1]
return sigma_1, sigma_2
# Function to compute alpha and k from friction angle and cohesion
def drucker_prager_parameters(friction_angle_deg, cohesion):
# Convert friction angle to radians
phi = np.radians(friction_angle_deg)
# Compute alpha and k
alpha = (2 * np.sin(phi)) / (np.sqrt(3) * (3 - np.sin(phi)))
# alpha = 0
k = (6 * cohesion * np.cos(phi)) / (np.sqrt(3) * (3 - np.sin(phi)))
return alpha, k
# Elasticity matrix (2D plane strain assumption)
def elasticity_matrix(E, nu):
D = (E / ((1 + nu) * (1 - 2 * nu))) * np.array([
[1 - nu, nu, 0],
[nu, 1 - nu, 0],
[0, 0, (1 - 2 * nu) / 2]
])
return D
# Invariants of the stress tensor
def compute_invariants(stress, stress_condition="plane_strain"):
sigma_xx, sigma_yy, tau_xy = stress
if stress_condition != "plane_strain":
# First invariant
I1 = sigma_xx + sigma_yy
# J2 invariant
# Deviatoric stresses
s_xx = sigma_xx - I1 / 2
s_yy = sigma_yy - I1 / 2
s_xy = tau_xy
# J2 = 0.5 * s:s
J2 = 0.5 * (s_xx**2 + s_yy**2 + s_xy**2)
# The above is equivalent to:
# J2 = (1/4) * (sigma_x - sigma_y)**2 + tau_xy**2
else: # Plain strain constraint
sigma_zz = nu * (sigma_xx + sigma_yy)
I1 = sigma_xx + sigma_yy + sigma_zz
# Deviatoric stresses
s_xx = sigma_xx - I1 / 3
s_yy = sigma_yy - I1 / 3
s_zz = sigma_zz - I1 / 3
s_xy = tau_xy
# J2 = 0.5 s:s
J2 = 0.5 * (s_xx**2 + s_yy**2 + s_zz**2 + s_xy**2)
return I1, J2
# Drucker-Prager yield function
def drucker_prager_yield(stress, alpha, k):
I1, J2 = compute_invariants(stress)
return alpha * I1 + np.sqrt(J2) - k
# Gradient of yield function df/dsigma
def df_dsigma(stress, alpha):
sigma_x, sigma_y, tau_xy = stress
I1, J2 = compute_invariants(stress)
dfdx = alpha + (sigma_x - sigma_y) / np.sqrt(J2)
dfdy = alpha - (sigma_x - sigma_y) / np.sqrt(J2)
dftau = 2 * tau_xy / np.sqrt(J2)
return np.array([dfdx, dfdy, dftau])
# Update stress and strain process
def update_stress_strain(stress_n, strain_n, strain_increment, D, alpha, k):
# Step 1: Compute trial stress
stress_trial = stress_n + np.dot(D, strain_increment)
# Step 2: Check yield condition
f_trial = drucker_prager_yield(stress_trial, alpha, k)
# print(f_trial)
if f_trial <= 0:
# Elastic step: no plastic deformation
return stress_trial, strain_n + strain_increment, 0.0 # No plastic multiplier
# Step 3: Plastic step - compute the plastic multiplier Δλ
df_sigma = df_dsigma(stress_trial, alpha)
H = np.dot(df_sigma, np.dot(D, df_sigma))
delta_lambda = f_trial / H
# Step 4: Update stress using the return mapping scheme
stress_updated = stress_trial - delta_lambda * np.dot(D, df_sigma)
# stress_updated[1] = 100 # yc: keep sigma_y constant for triaxial test
# Step 5: Update plastic strain
plastic_strain_increment = delta_lambda * df_sigma
strain_updated = strain_n + plastic_strain_increment
return stress_updated, strain_updated, delta_lambda
Material properties#
# Material properties
E = 100000
nu = 0.3
rho = 3600
mu = 0.385
phi = 30
c = 1
K0 = 0.5
Analysis setting#
# Analysis setting
dtime = 0.0001
time = 1.0
t = 0
interval = 200
g = 4.8
Mesh generation#
# length of domain
l = 1
# number of elements for each dim
n_ele_x = 20
n_ele_y = 20
# cell size
delta_x = l / n_ele_x
delta_y = l / n_ele_y
# number of grid nodes
n_node_x = n_ele_x + 1
n_node_y = n_ele_y + 1
n_nodes = n_node_x * n_node_y
# Generate mesh nodes
x_coords = np.linspace(0, l, n_node_x)
y_coords = np.linspace(0, l, n_node_y)
X, Y = np.meshgrid(x_coords, y_coords)
nodes = np.vstack([X.flatten(), Y.flatten()]).T
# Generate elements (connectivity)
elements = []
for j in range(n_node_y - 1):
for i in range(n_node_x - 1):
n1 = j * n_node_x + i
n2 = n1 + 1
n3 = n2 + n_node_x
n4 = n1 + n_node_x
elements.append([n1, n2, n3, n4])
elements = np.array(elements)
elements_coordinate_map = nodes[elements]
n_elements = elements.shape[0]
# Boundary nodes
bottomNodes = np.where(nodes[:, 1] < 1e-8)[0]
upperNodes = np.where(nodes[:, 1] > l - 1e-8)[0]
leftNodes = np.where(nodes[:, 0] < 1e-8)[0]
rightNodes = np.where(nodes[:, 0] > l - 1e-8)[0]
# Initialize node quantities
n_masses = np.zeros(n_nodes) # nodal masses
n_momentums = np.zeros((n_nodes, 2)) # nodal momentums
n_iforces = np.zeros((n_nodes, 2)) # Internal forces
n_eforces = np.zeros((n_nodes, 2)) # External forces. Not used in this practice
# Initialize grid state variables
gStress = np.zeros((3, n_nodes)) # [sig_xx, sig_yy, sig_xy]
gDisp = np.zeros((2, n_nodes)) # [disp_x, disp_y]
Generate particles#
# Inputs for particles representing granular column
origin = [0.0125, 0.0125]
lengths = [0.300, 0.300]
n_particle_per_cell_per_dim = 2
# Generate particles for granular column body
spacing = delta_x / n_particle_per_cell_per_dim
xp = generate_rectangular_particles(
origin=origin, length=lengths[0], height=lengths[1], spacing=spacing)
# particle states
vp = np.zeros(xp.shape)
s = np.zeros((len(xp), 3)) # [sigma_xx, sigma_yy, sigma_xy]
eps = np.zeros((len(xp), 3)) # [epsilon_xx, epsilon_yy, gamma_xy]
Fp = np.tile([1.0, 0.0, 0.0, 1.0], (len(xp), 1))
# Find elements to which particles belong
ele_ids_of_particles, p_ids_in_eles = particle_element_mapping(
xp, delta_x, delta_y, n_ele_x, n_elements)
active_elements = np.unique(ele_ids_of_particles)
active_nodes = np.unique(elements[active_elements, :])
# Compute initial particle volume
Vp = np.zeros(len(xp))
# Volume (area) of each background cell
for p_ids_in_ele in p_ids_in_eles:
n_mp_in_element = len(p_ids_in_ele)
if n_mp_in_element > 0:
volume_per_mp = (delta_x * delta_y) / n_mp_in_element
Vp[p_ids_in_ele] = volume_per_mp
Vp0 = Vp.copy() # save initial volume
Mp = Vp * rho # mass of particles
# Initialize stress
height = lengths[1] + spacing / 2
s[:, 1] = - g * rho * (height - xp[:, 1])
s[:, 0] = K0 * s[:, 1]
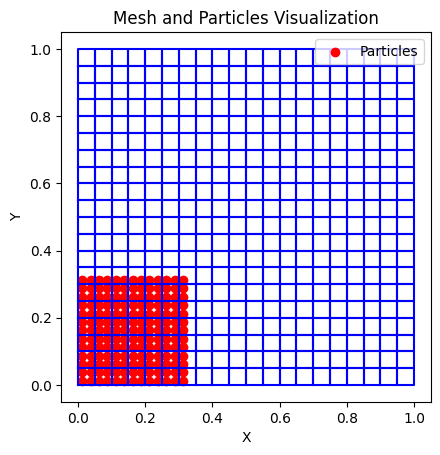
# Plot mesh, particles
meshes = [(nodes, elements)]
def plot_mesh_and_particles(nodes, elements, particles):
fig, ax = plt.subplots()
# Plot the mesh
for element in elements:
# Get the coordinates of the element's nodes
quad_coords = nodes[element]
# Repeat the first point to close the quad
quad_coords = np.vstack([quad_coords, quad_coords[0]])
ax.plot(quad_coords[:, 0], quad_coords[:, 1], 'b-', linewidth=1.5) # Blue lines for the mesh
# Plot the particles
ax.scatter(particles[:, 0], particles[:, 1], color='red', marker='o', label='Particles')
# Set labels and title
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title('Mesh and Particles Visualization')
ax.legend()
# Equal aspect ratio
ax.set_aspect('equal', 'box')
plt.show()
plot_mesh_and_particles(nodes, elements, xp)
Run analysis#
# Results to save
ta = [] # time
ka = [] # kinetic energy
pa = [] # potential energy
da = [] # dissipation energy
# sa = [] # strain energy
pos = [] # positions
vel = [] # velocities
stresses = []
strains = []
nsteps = int(time / dtime)
istep = 0
potential_e_t0 = np.sum(Mp * g * xp[:, 1])
for istep in tqdm(range(nsteps), desc="Simulating"):
# print(f"Time: {t:.5f}/{time}")
# Compute potential energy
potential_e = np.sum(Mp * g * xp[:, 1])
kinetic_e = np.sum(0.5 * Mp * np.linalg.norm(vp, axis=1)**2)
dissipation_e = potential_e_t0 - kinetic_e - potential_e
# Refresh the nodal values
n_masses.fill(0)
n_momentums.fill(0)
n_iforces.fill(0)
n_eforces.fill(0)
gStress.fill(0)
# Iterate over the computational cells (i.e., elements)
for ele_id in active_elements:
# node ids of the current element
node_ids = elements[ele_id]
# coords of the current element
node_coords = nodes[node_ids, :]
# particle ids inside the current element
material_points = p_ids_in_eles[ele_id]
# Iterate over the material points in the current cell
for p in material_points:
# Convert global coordinate "p=(x, y)" to local coordiate "pt=(xi, eta)".
pt = np.array(
[(2 * xp[p, 0] - (node_coords[0, 0] + node_coords[1, 0])) / delta_x,
(2 * xp[p, 1] - (node_coords[1, 1] + node_coords[2, 1])) / delta_y])
# Evaluate shape function and its derivatives with respect to local coords (xi, eta) at (x, y).
N, dNdxi = lagrange_basis("Q4", pt)
# Evaluate the Jacobian at current the current local coords (xi, eta).
J0 = node_coords.T @ dNdxi
# Get the inverse of Jacobian
invJ0 = np.linalg.inv(J0)
# Get the derivative of shape function with respect to global coords, i.e., (x, y)
dNdx = dNdxi @ invJ0
# Current stress of material points
stress = s[p, :]
# Iterate over the nodes of the current element & update nodal values by interpolating material point values
for i, node_id in enumerate(node_ids):
dNIdx = dNdx[i, 0]
dNIdy = dNdx[i, 1]
n_masses[node_id] += N[i] * Mp[p]
n_momentums[node_id, :] += N[i] * Mp[p] * vp[p, :]
n_iforces[node_id, 0] -= Vp[p] * (stress[0] * dNIdx + stress[2] * dNIdy)
n_iforces[node_id, 1] -= Vp[p] * (stress[2] * dNIdx + stress[1] * dNIdy)
n_eforces[node_id, 1] -= N[i] * Mp[p] * g
# Total nodal force
nforce = n_iforces + n_eforces
# Compute nodal accelerations
n_accelerations = np.zeros((n_nodes, 2))
# valid_mass = n_masses > 1e-12
# n_accelerations[valid_mass, :] = nforce[valid_mass, :] / n_masses[valid_mass, None]
n_accelerations[active_nodes, :] = nforce[active_nodes, :] / n_masses[active_nodes, None]
# Boundary conditions
# 0. Update nodal kinematics (nodal mass, momentum -> particles)
# 1. Velocity & acceleration to 0
# 2. Compute nodal force.
# * First, map particle body force to nodal body force
# * Next, map particle internal force to nodel internal force
# 3. compute_particle_kinematics.
# * compute_acceleration_velocity
# * accel = (f_e + f_i) / mass
# * apply_friction_constraints
# * vel += accel * dt
# * apply_velocity_constraints
# * compute_updated_position
# Function to apply frictional boundary condition on given nodes
def apply_frictional_boundary_condition(node_ids, dir_n, sign_dir_n):
dir_t = 1 - dir_n # Tangential direction
# Normal and tangential accelerations
acc_n = n_accelerations[node_ids, dir_n]
acc_t = n_accelerations[node_ids, dir_t]
# Tangential velocities
vel_t = n_momentums[node_ids, dir_t] / n_masses[node_ids]
# Determine nodes where particles are acting towards the boundary
positive_movement_towards_boundary = (acc_n * sign_dir_n) > 0.0
# Apply frictional boundary condition
for idx in np.where(positive_movement_towards_boundary)[0]:
node_id = node_ids[idx]
acc_n_i = acc_n[idx]
acc_t_i = acc_t[idx]
vel_t_i = vel_t[idx]
# print(vel_t_i)
# Determine static or kinetic friction
if vel_t_i != 0.0: # Kinetic friction
# compute tangential velocity at next timestep
vel_net = dtime * acc_t_i + vel_t_i
vel_frictional = dtime * mu * abs(acc_n_i)
if abs(vel_net) <= vel_frictional:
# friction stops the particle
acc_t_i = -vel_t_i / dtime
else:
# friction reduces the tangential acceleration
acc_t_i -= np.sign(vel_net) * mu * abs(acc_n_i)
# acc_t_i -= np.sign(vel_net) * mu * abs(acc_n_i)
else: # Static friction
if abs(acc_t_i) <= mu * abs(acc_n_i):
acc_t_i = 0.0
else:
acc_t_i -= np.sign(acc_t_i) * mu * abs(acc_n_i)
# Update tangential acceleration
n_accelerations[node_id, dir_t] = acc_t_i
# Update nodal force at this node
nforce[node_id, :] = n_accelerations[node_id, :] * n_masses[node_id]
apply_frictional_boundary_condition(
np.intersect1d(leftNodes, active_nodes), dir_n=0, sign_dir_n=-1.0)
apply_frictional_boundary_condition(
np.intersect1d(rightNodes, active_nodes), dir_n=0, sign_dir_n=1.0)
apply_frictional_boundary_condition(
np.intersect1d(bottomNodes, active_nodes), dir_n=1, sign_dir_n=-1.0)
apply_frictional_boundary_condition(
np.intersect1d(upperNodes, active_nodes), dir_n=1, sign_dir_n=1.0)
# Impose boundary condition
n_momentums[leftNodes, 0] = 0
n_momentums[rightNodes, 0] = 0
n_momentums[bottomNodes, 1] = 0
n_momentums[upperNodes, 1] = 0
nforce[leftNodes, 0] = 0
nforce[rightNodes, 0] = 0
nforce[bottomNodes, 1] = 0
nforce[upperNodes, 1] = 0
# Update nomal momentum
n_momentums += nforce * dtime
k = 0
u = 0
# Iterate over the computational cells (i.e., elements)
for ele_id in active_elements:
# node ids of the current element
node_ids = elements[ele_id]
# coords of the current element
node_coords = nodes[node_ids, :]
# particle ids inside the current element
material_points = p_ids_in_eles[ele_id]
# Iterate over the material points in the current cell
for p in material_points:
# Convert global coordinate "p=(x, y)" to local coordiate "pt=(xi, eta)".
pt = np.array(
[(2 * xp[p, 0] - (node_coords[0, 0] + node_coords[1, 0])) / delta_x,
(2 * xp[p, 1] - (node_coords[1, 1] + node_coords[2, 1])) / delta_y])
N, dNdxi = lagrange_basis("Q4", pt)
J0 = node_coords.T @ dNdxi
invJ0 = np.linalg.inv(J0)
dNdx = dNdxi @ invJ0
Lp = np.zeros((2, 2))
for i, node_id in enumerate(node_ids):
vI = np.zeros(2)
vp[p, :] += dtime * N[i] * nforce[node_id, :] / n_masses[node_id]
xp[p, :] += dtime * N[i] * n_momentums[node_id, :] / n_masses[node_id]
vI = n_momentums[node_id, :] / n_masses[node_id] # nodal velocity
Lp += vI.reshape(2, 1) @ dNdx[i, :].reshape(1, 2) # particle velocity gradient
F = Fp[p, :].reshape(2, 2) @ (np.eye(2) + Lp * dtime)
Fp[p, :] = F.flatten()
Vp[p] = np.linalg.det(F) * Vp0[p]
dEps = dtime * 0.5 * (Lp + Lp.T)
D = elasticity_matrix(E, nu)
alpha, k = drucker_prager_parameters(phi, c)
new_sig, new_epsE, _ = update_stress_strain(
stress_n=s[p, :],
strain_n=eps[p, :],
strain_increment=np.array([dEps[0, 0], dEps[1, 1], dEps[0, 1]]),
D=D,
alpha=alpha,
k=k)
s[p, :] = new_sig
eps[p, :] = new_epsE
for i, node_id in enumerate(node_ids):
gStress[:, node_id] += N[i] * s[p, :]
stresses.append(s.copy())
strains.append(eps.copy())
pos.append(xp.copy())
vel.append(vp.copy())
# Update particle-element mapping (elements to which particles belong)
ele_ids_of_particles, p_ids_in_eles = particle_element_mapping(
xp, delta_x, delta_y, n_ele_x, n_elements)
active_elements = np.unique(ele_ids_of_particles)
active_nodes = np.unique(elements[active_elements, :])
if istep % interval == 0:
# result = {"positions": xp, "stress": s}
# save_dir = f"./results_vtk_column/"
# visualizer.save_vtk(istep, result, save_dir)
ta.append(t)
ka.append(kinetic_e)
pa.append(potential_e)
da.append(dissipation_e)
t += dtime
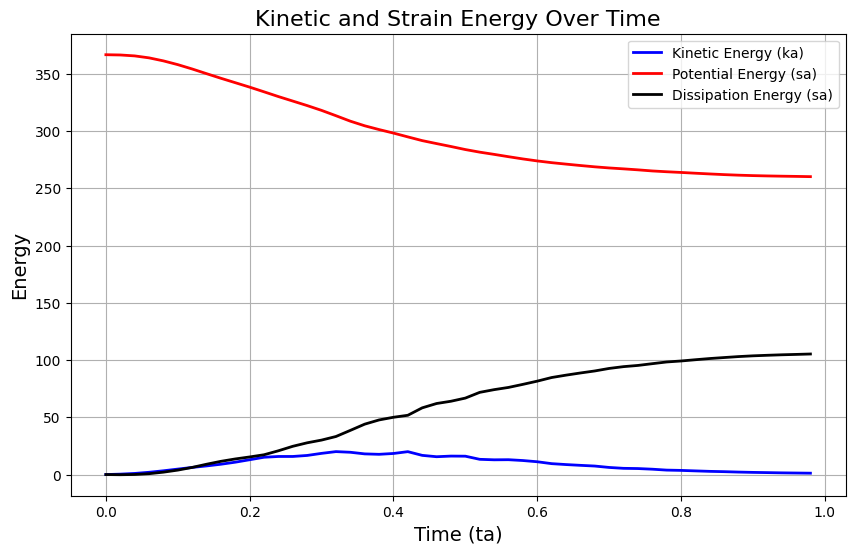
# Plot kinetic and strain energy
plt.figure(figsize=(10, 6))
plt.plot(ta, ka, label='Kinetic Energy (ka)', color='b', linewidth=2)
plt.plot(ta, pa, label='Potential Energy (sa)', color='r', linewidth=2)
plt.plot(ta, da, label='Dissipation Energy (sa)', color='k', linewidth=2)
plt.xlabel('Time (ta)', fontsize=14)
plt.ylabel('Energy', fontsize=14)
plt.title('Kinetic and Strain Energy Over Time', fontsize=16)
plt.legend()
plt.grid(True)
plt.show()
Simulating: 100%|██████████| 10000/10000 [03:12<00:00, 52.00it/s]
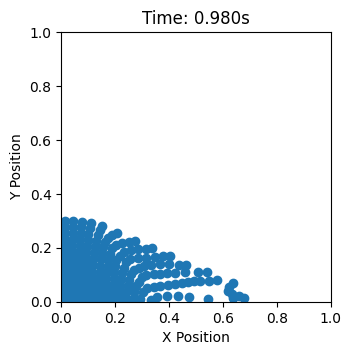
Visualize trajectory#
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
# Create figure and axis
fig, ax = plt.subplots(figsize=(5, 3.5))
ax.set_xlim(0, l)
ax.set_ylim(0, l)
ax.set_xlabel('X Position')
ax.set_ylabel('Y Position')
ax.set_title('Granular Column Collapse')
# Initialize scatter plot
scatter = ax.scatter([], [])
# Animation update function
def update(frame):
scatter.set_offsets(pos[frame * interval])
ax.set_title(f'Time: {ta[frame]:.3f}s')
ax.set_aspect('equal')
return scatter,
# Create animation
# Using the same interval as your save interval
anim = FuncAnimation(
fig, update,
frames=len(ta), # number of frames matches saved timesteps
interval=50, # 50ms between frames
blit=True
)
# Display animation in notebook
HTML(anim.to_jshtml())
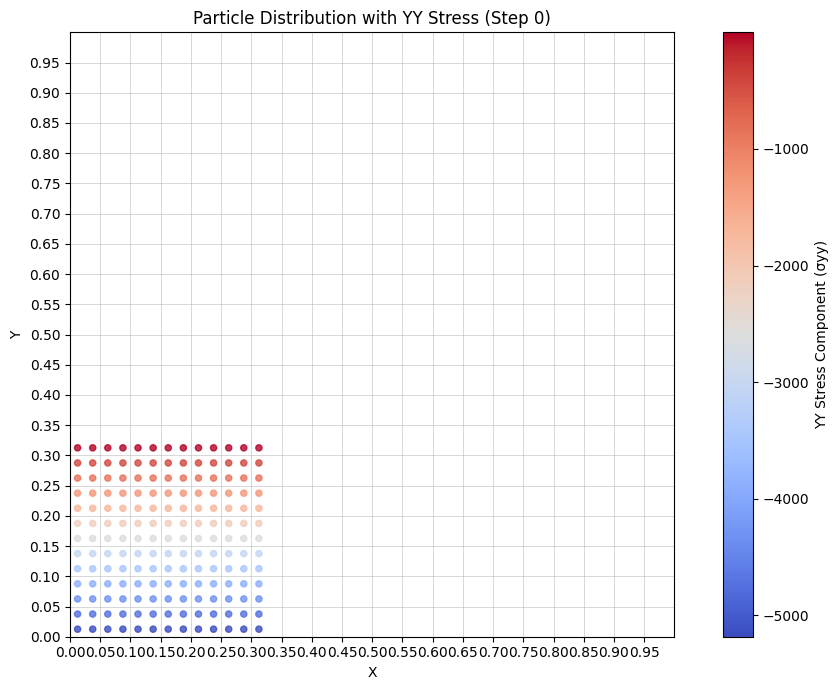
Visualize stress#
# Create animation of particles colored by yy stress
fig, ax = plt.subplots(figsize=(10, 7))
# Set axis properties
ax.set_aspect('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title('Particle Distribution with YY Stress')
ax.grid(True)
# Set fixed axis limits based on domain size
ax.set_xlim(0, l)
ax.set_ylim(0, l)
# Create a normalized colormap - use consistent limits for better visualization
vmin = min([np.min(stresses[0][:, 1]) for i in range(len(stresses))])
vmax = max([np.max(stresses[0][:, 1]) for i in range(len(stresses))])
# Create a ScalarMappable for the colorbar outside the animation function
sm = plt.cm.ScalarMappable(cmap='coolwarm', norm=plt.Normalize(vmin=vmin, vmax=vmax))
sm.set_array([]) # You need to set an array (empty in this case)
cbar = plt.colorbar(sm, ax=ax)
cbar.set_label('YY Stress Component (σyy)')
def update_stress_plot(frame):
# Clear previous frame
ax.clear()
# Get yy stress component (s[:, 1]) for current frame
yy_stress = stresses[frame * interval][:, 1]
# Plot particles colored by stress
scatter = ax.scatter(
pos[frame * interval][:, 0],
pos[frame * interval][:, 1],
c=yy_stress,
cmap='coolwarm',
vmin=vmin,
vmax=vmax,
s=20,
alpha=0.8
)
# Set axis properties
ax.set_aspect('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title(f'Particle Distribution with YY Stress (Step {frame * interval})')
ax.grid(True)
ax.grid(True, linestyle='-', linewidth=0.5, alpha=0.7)
ax.set_xticks(np.arange(0, l, l/n_ele_x))
ax.set_yticks(np.arange(0, l, l/n_ele_y))
# Set fixed axis limits based on domain size
ax.set_xlim(0, l)
ax.set_ylim(0, l)
plt.tight_layout()
return scatter,
# Create animation
from matplotlib.animation import FuncAnimation
anim = FuncAnimation(
fig,
update_stress_plot,
frames=len(ta),
interval=100, # milliseconds between frames
blit=False
)
# Display the animation in the notebook
from IPython.display import HTML
HTML(anim.to_jshtml())
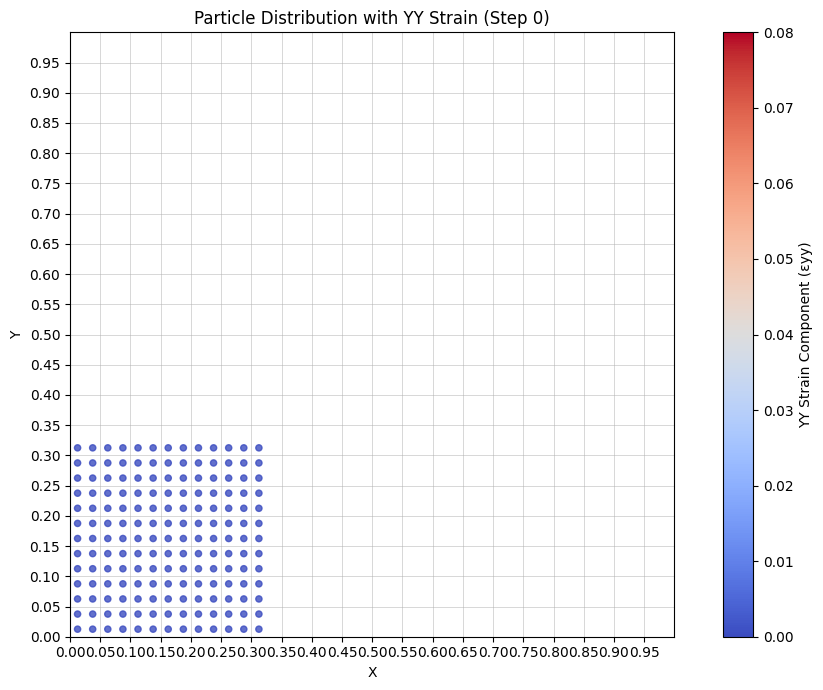
Visualize strain#
# Create animation of particles colored by yy strain
fig, ax = plt.subplots(figsize=(10, 7))
# Set axis properties
ax.set_aspect('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title('Particle Distribution with YY Strain')
ax.grid(True)
# Set fixed axis limits based on domain size
ax.set_xlim(0, l)
ax.set_ylim(0, l)
# Create a normalized colormap - use consistent limits for better visualization
vmin = 0
vmax = 0.08
# Create a ScalarMappable for the colorbar outside the animation function
sm = plt.cm.ScalarMappable(cmap='coolwarm', norm=plt.Normalize(vmin=vmin, vmax=vmax))
sm.set_array([]) # You need to set an array (empty in this case)
cbar = plt.colorbar(sm, ax=ax)
cbar.set_label('YY Strain Component (εyy)')
def update_strain_plot(frame):
# Clear previous frame
ax.clear()
# Get yy strain component (eps[:, 1]) for current frame
xy_strain = strains[frame * interval][:, 2]
# Plot particles colored by strain
scatter = ax.scatter(
pos[frame * interval][:, 0],
pos[frame * interval][:, 1],
c=xy_strain,
cmap='coolwarm',
vmin=vmin,
vmax=vmax,
s=20,
alpha=0.8
)
# Set axis properties
ax.set_aspect('equal')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.set_title(f'Particle Distribution with YY Strain (Step {frame * interval})')
ax.grid(True)
ax.grid(True, linestyle='-', linewidth=0.5, alpha=0.7)
ax.set_xticks(np.arange(0, l, l/n_ele_x))
ax.set_yticks(np.arange(0, l, l/n_ele_y))
# Set fixed axis limits based on domain size
ax.set_xlim(0, l)
ax.set_ylim(0, l)
plt.tight_layout()
return scatter,
# Create animation
from matplotlib.animation import FuncAnimation
strain_anim = FuncAnimation(
fig,
update_strain_plot,
frames=len(ta),
interval=100, # milliseconds between frames
blit=False
)
# Display the animation in the notebook
from IPython.display import HTML
HTML(strain_anim.to_jshtml())